Jean Hausser Lab
Quantitative tumor immune control

Research field
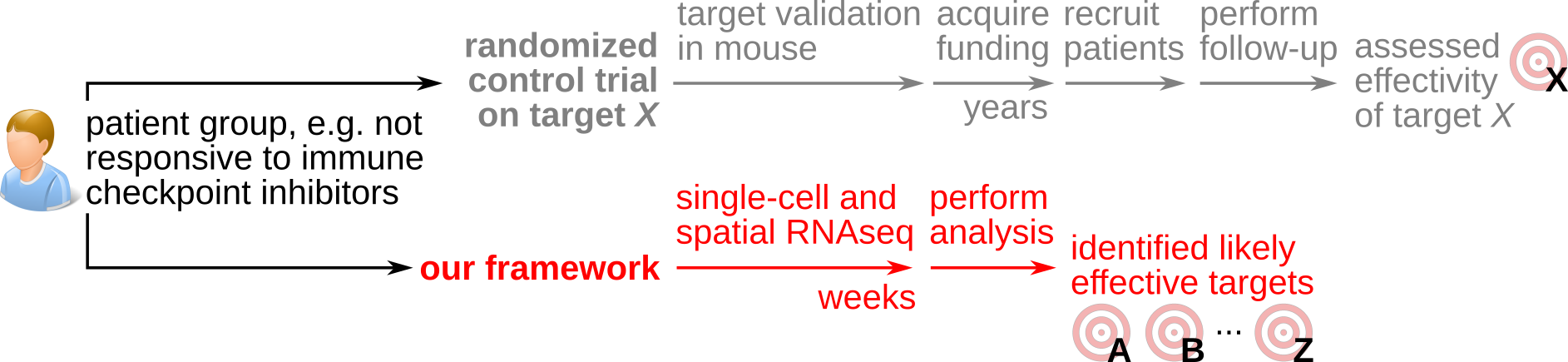
Immunotherapy has transformative potential in cancer, as illustrated by the durable remission of ~30% of metastatic patients with immune checkpoint blockade therapy in several cancer types. Yet, most patients do not respond to immune checkpoint blockade. Engineering effective immunotherapy is challenging due to the complexity of tumor immunology as a biosystem, with dozens of cell types whose activity is potentially controlled by hundreds of molecular signals and thousands of genes.
Historically, controlling and engineering complex systems - from electronic circuits to space vessels - has benefited from articulating these systems into quantitative theories. A quantitative theory of tumor immunology is presently missing. Formulating such a theory can be catalyzed by the growing tumor single-cell and spatial omics data which can quantify thousands of signals and genes in the local contexts of human tumors. We are still missing, however, a quantitative framework to turn this data into predictive models of how signals and genes determine tumor fate.
Approach and vision
To address this, we are researching a quantitative framework of tumor immune control designed to maximize the utility of human tumor spatial and single-cell transcriptomics data for engineering personalized immunotherapy. We take inspiration from physics-style mathematical modeling which we implement into new data science and machine-learning methods. We calibrate and validate these through experiments in vitro and in vivo.
Our vision is for our quantitative framework to accelerate immunotherapeutic innovation, by identifying targets from patient material within months or even weeks, instead of the many years needed with current target identification approaches.
Selected publications
Full list of our publications on Google Scholar.
Multicellular tumor-immune dynamics reveal therapeutic windows
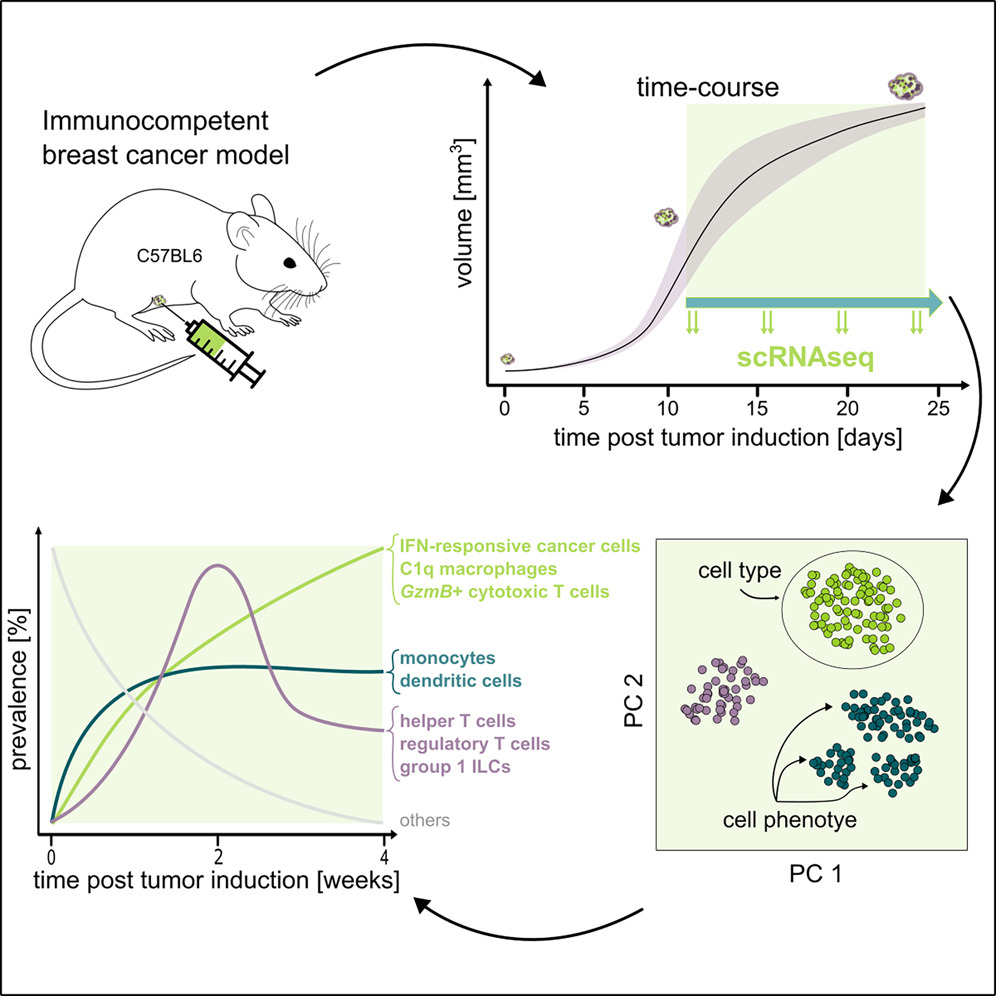
Tumors are complex ecosystems where cancer cells, immune cells, and stromal cells interact dynamically. Yet most studies profile tumors at single timepoints, capturing static snapshots rather than the temporal choreography of how tumors build their microenvironment. This prevents us from distinguishing causes from consequences and from identifying when each cell type arrives and what it does – critical for timing therapeutic interventions.
We performed a longitudinal single-cell RNA-sequencing study to film the movie of tumor progression of in a mouse breast cancer model. We found that tumor microenvironment dynamics follow three distinct temporal patterns: stable colonization (monocytes, dendritic cells establish early), wave-like dynamics (helper and regulatory T cells surge at week 3 then recede), and progressive increase (interferon-responsive cancer cells and immunosuppressive C1q+ macrophages accumulate continuously).
These patterns reveal therapeutic opportunities: the week-3 T cell surge could represent a potential intervention window; blocking interferon-response pathways could strip cancer cells of their immune armor; and reprogramming C1q+ macrophages from immunosuppressive to anti-tumor states could restore immune control. We also found that genetically identical tumors progress at different speeds – some reach advanced immune states in 11 days, others in 18 days – uncorrelated with tumor volume or growth rate, revealing an unexplained tumor clock that likely reflects individual host factors. This establishes a framework for understanding how cell-cell interactions evolve temporally in disease.
Multi-cellular phenotypic dynamics during the progression of an immunocompetent breast cancer model, Louise Gsell, Spencer S. Watson, Iva Sutevski, Matteo Massara, Klara Soukup, Alper Eroglu, Jeff E. Mold, Antony Cougnoux, Johanna A. Joyce, Jean Hausser. iScience 2025
Niche-phenotype mapping (NIPMAP) uses ecological niche theory to reveal hidden structure in spatial tissue data
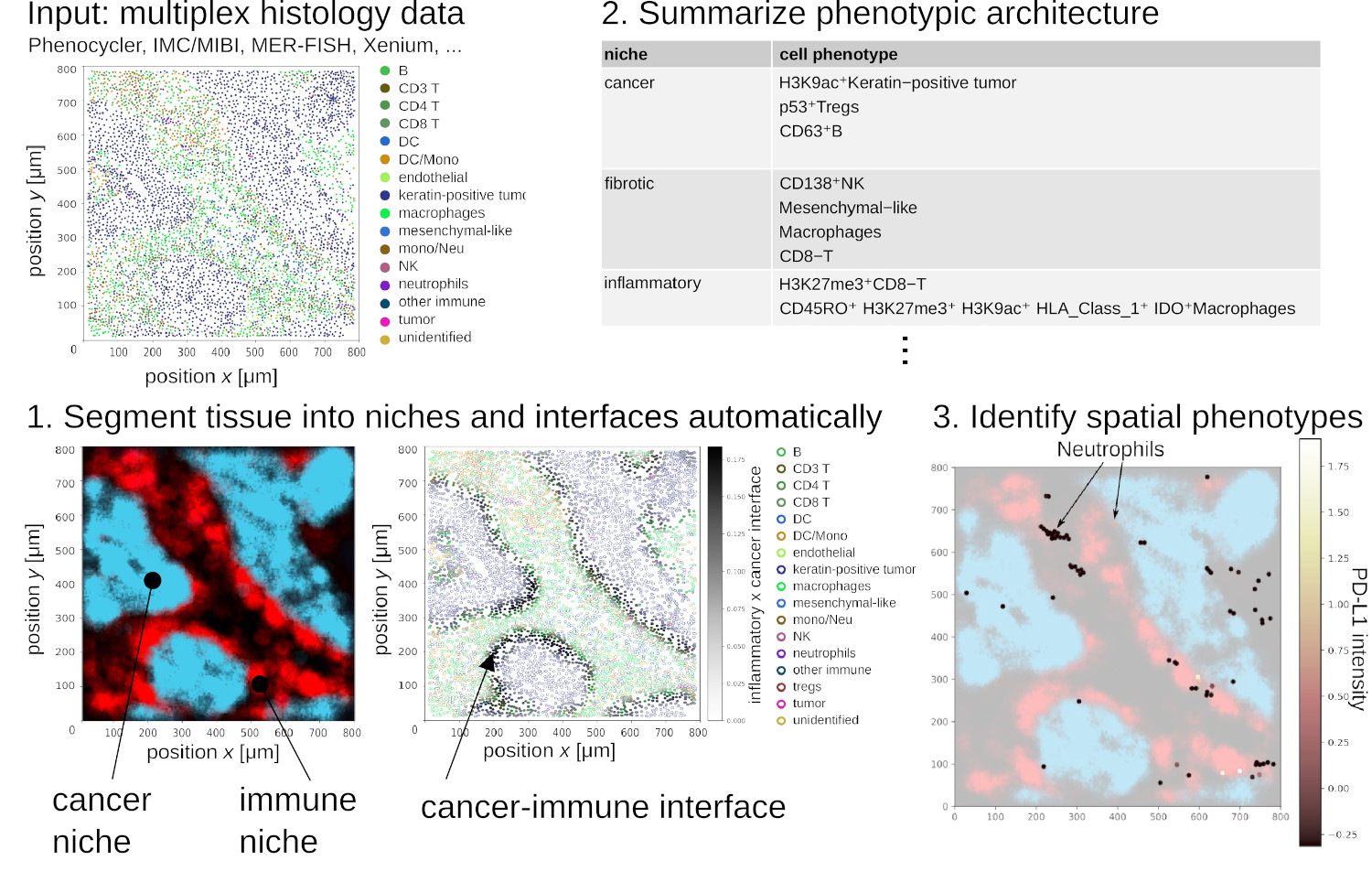
The spatial architecture of tumors has high relevance for diagnostic and therapy and can be surveyed by multiplex histology techniques such as imaging mass cytometry and multiplex immunofluorescence. This produces spatial maps of dozens to hundreds of cellular and phenotypic markers. But surveying these spatial maps exhaustively requires visualizing manually 10'000+ images per sample. To address this, NIPMAP uses unsupervised machine-learning to (1) concisely and accurately summarize the architecture of tissues, and (2) automatically identify phenotypes with salient spatial architecture.
We are presently leveraging NIPMAP for precision oncology, in collaboration with two clinical labs in France and USA.
NIPMAP: niche-phenotype mapping of multiplex histology data by community ecology, Anissa El Marrahi, Fabio Lipreri, Ziqi Kang, Louise Gsell, Alper Eroglu, David Alber, Jean Hausser. Nature Communications 2023
Multi-task tumor evolution
Previously, we researched a quantitative framework to explain the diversity of gene expression, mutations and drug sensitivities of tumors, grounded in evolutionary theory. This framework suggests approaches to overcome cancer diversity, which represents a major challenge to therapy.
Universal cancer tasks, evolutionary trade-offs, and the function of driver mutations. Jean Hausser, Pablo Szekely, Noam Bar, Hila Sheftel, Carlos Caldas, Uri Alon. Nature Communications 2019.
 Tumor
heterogeneity and the evolutionary trade-offs of
cancer. Jean Hausser, Uri Alon. Nature Reviews
Cancer 2020.
Tumor
heterogeneity and the evolutionary trade-offs of
cancer. Jean Hausser, Uri Alon. Nature Reviews
Cancer 2020.
We demonstrated that this framework can guide the design of drug combinations effective against heterogeneous populations of cancer cells using 3D tumor spheroids.
Microscopy-based phenotypic monitoring of MDA-MB-231 spheroids allows the evaluation of phenotype-directed therapy. Loay Mahmoud, Antony Cougnoux, Christina Bekiari, Paloma Araceli Ruiz de Castroviejo Teba, Anissa El Marrahi, Guilhem Panneau, Louise Gsell, Jean Hausser. Experimental Cell Research 2023.
Team
 Jean Hausser
Jean Hausser
principal investigator (CV)
jean.hausser@scilifelab.se
 Sampath Narayanan
Sampath Narayanan
staff scientist (experimental biology)
sampath.narayanan@scilifelab.se
 Shibani Veeraragavan
Shibani Veeraragavan
post-doctoral fellow (biomathematics)
shibani.veeraragavan@scilifelab.se
 Alper Eroglu
Alper Eroglu
PhD student (computational biology)
alper.eroglu@scilifelab.se
 Louise Gsell
Louise Gsell
PhD student (computational biology)
louise.gsell@scilifelab.se
 Chenxi Sun
Chenxi Sun
Master thesis student (biomedicine)
 Egle Fissore
Egle Fissore
Master thesis student (precision medicine)
Alumni
 Henrik Palmér (2025)
Henrik Palmér (2025)
master thesis student
position after the lab: medical student, Karolinska Institutet
 Jorrit Stada (2025)
Jorrit Stada (2025)
bachelor thesis student (molecular life science)
position after the lab: PhD student, Karolinska Institute
 Emma Falco (2025)
Emma Falco (2025)
bachelor thesis student (cell biology)
position after the lab: masters in immunology, Grenoble France
 Chuhanwen Sun
Chuhanwen Sun
master student (2024-2025)
position after the lab: PhD student, Rutgers University
 Dongxiao Chen (2024)
Dongxiao Chen (2024)
master student (bioinformatics)
position after the lab: data science in industry, Nanjing China
 Antony Cougnoux (2020-2023)
Antony Cougnoux (2020-2023)
staff scientist (immunology, cancer)
position after the lab: principal investigator, NIH Bethesda
 Jakob Rosenbauer (2021-2023)
Jakob Rosenbauer (2021-2023)
EMBO post-doctoral fellow (physics)
position after the lab: postdoc, DKFZ Heidelberg
 Mathilda Stigenberg (2023)
Mathilda Stigenberg (2023)
master student (bioinformatics)
position after the lab: research assistant, Uppsala University
 Felix Waern (2023)
Felix Waern (2023)
master student (bioinformatics)
position after the lab: PhD student, Karolinska Institutet
 Sofie Blahova (2023)
Sofie Blahova (2023)
master student (bioengineering)
position after the lab: PhD student, Karolinska Institutet
 Anissa El Marrahi (2020-2023)
Anissa El Marrahi (2020-2023)
master thesis student, research assistant, bioinformatician
position after the lab: PhD student, IRB Bellinzona
 Raziyeh Mohseni (2023)
Raziyeh Mohseni (2023)
bachelor student (bioinformatics)
position after the lab: bioinformatics masters program Uppsala University
 Petter Säterskog (2021-2023)
Petter Säterskog (2021-2023)
post-doctoral fellow (theoretical physics)
 Loay Mahmoud (2021-2023)
Loay Mahmoud (2021-2023)
research assistant (experimental biomedicine)
position after the lab: PhD student, NTNU Trondheim
 Ziqi Kang (2022-2023)
Ziqi Kang (2022-2023)
master thesis student, research assistant (bioinformatics)
position after the lab: PhD student, Helsinki University
 Iva Sutevski (2022-2023)
Iva Sutevski (2022-2023)
research assistant (biology)
position after the lab: PhD student, KTH Stockholm
 Benjamin Maier (2021-2022)
Benjamin Maier (2021-2022)
visiting student (molecular life science)
position after the lab: PhD student, EMBL Heidelberg
 Javier Escudero Morlanes (2021-2022)
Javier Escudero Morlanes (2021-2022)
master student (molecular life science)
position after the lab: research engineer, KTH/SciLifeLab Stockholm
 Christina Bekiari (2022)
Christina Bekiari (2022)
master student (biomedicine)
position after the lab: research assistant, Stockholm University/SciLifeLab
 Guilhem Panneau (2021-2022)
Guilhem Panneau (2021-2022)
research assistant (computational biology)
position after the lab: PhD student, University of Lausanne
 Paloma Araceli Ruiz de Castroviejo Teba (2021)
Paloma Araceli Ruiz de Castroviejo Teba (2021)
visiting student (biomedicine)
position after the lab: product development scientist, Basic Genomics Stockholm
 Fabio Lipreri (2020-2021)
Fabio Lipreri (2020-2021)
research assistant (computer science)
position after the lab: data scientist, FrescoFrigo Milano
 Letizia Orsini (2020-2021)
Letizia Orsini (2020-2021)
research assistant (biostatistics)
position after the lab: biostatistician, Karolinska Institutet
 Axel de Tonnac (2020-2021)
Axel de Tonnac (2020-2021)
consultant (bioengineering / bioinformatics)
position after the lab: sales executive, Sophia Genetics Lausanne
 Tagore Sanketh Bandaru (2020)
Tagore Sanketh Bandaru (2020)
master thesis student (molecular life science)
position after the lab: PhD student, University Hospital Basel
 Maximilian Reck (2020)
Maximilian Reck (2020)
master thesis student (molecular life science)
position after the lab: PhD student, University of Edinburgh
 Fredrik Carlsson (2020)
Fredrik Carlsson (2020)
summer student and developper
position after the lab: medical student, Karolinska Institutet
 David Alber (2020)
David Alber (2020)
research student (computational physics)
position after the lab: data scientist, IBM Vienna
 Panos Kalogeropoulos (2020)
Panos Kalogeropoulos (2020)
summer student (molecular life science)
position after the lab: PhD student, Stockholm University
Joining us
- A postdoctoral fellow Email Jean about 1. which of your research project was most exciting to you so far and why, 2. why you'd like to join us (1000 words max) and 3. attach a concise CV.
- Master students enrolled in biology /
biomedical programs or computational
programs (bioinformatics, physics, applied
mathematics, computer science, engineering,
...).
Email Jean about 1. what course or projects you most enjoyed in your studies so far, 2. why you'd like to join us (1000 words maximum), and 3. attach a concise CV.
How to find us

The lab is located at Karolinska Institutet and SciLifeLab in Stockholm, Sweden. Within Karolinska Institutet, we are affiliated to the department of Cell and Molecular Biology.
Visiting address
Hausser lab
Biomedicum B6
Solnavägen 9, Solna
Mailing address
Hausser lab
Department of Cell and Molecular Biology (CMB)
Karolinska Institutet
P.O. Box 285
SE-171 77 Stockholm, Sweden
Telephone switch board: +46 (0) 8 524 800 00